Friends and denizens of the internet – I return to regale you with stories from inside the lab. To remind old readers and acquaint new ones, I seek here to peel the skin off the lab (ouch!). I mean to show in sort of real time how experiments are run, complete with broken test-tubes, equipment malfeasance, and all the rest of daily doings that fill up a scientist’s life but that never make the papers. To be sure my status as no-longer-larking-about-on-sabbatical means that I shall have far less time for anything inside the lab, so these posts probably won’t be every week. I’ll do my best.
Fair warning—this post is a bit longish because it spans many weeks of activity since my plane landed.
Last season’s finale, hardly a cliff-hanger, summarized the two experimental strands that I worked on at CPIB. Moving forward here at UMass Amherst, I plan to focus on just one – the root oscillation. While data from CPIB remain to be crunched and high-falootin’ numerical analyses remain to applied, there are experiments needing to be done. For one thing, does the oscillation depend on growth of the root inside the agar medium? Or will roots grown on the surface of the agar also oscillate? For another, does the oscillation reflect an oscillatory ‘something’ moving down from the shoot? Or will roots with their shoots cut off nevertheless oscillate?
Answering these questions should require fewer roots (and hence less crunching, yay!) than establishing the oscillation in the first place. This is because I will be seeing whether the oscillation is present (yes or no) rather than quantifying a changed parameter (e.g., period or amplitude).
The good news is that I have a sideways microscope (figure 1).

The bad news is that the camera is not abundantly supplied with pixels. Anticipating this, I brought back from the UK a camera with a 2400 pixel x 2000 pixel chip. This many pixels allows most of the growth zone to be in the field of view at a high enough pixel density to let the software calculate an accurate velocity profile. But this camera (an IDS µEye) has drivers for Windows (or Linux) computers only and my lab runs largely on the Mac side. Thus, the first task was to get an emulator going on the Mac and teach it run the µEye. After some hilarity trying to do this with Linux, I succeeded with Windows. Fortunately, the open source program Micro-Manager can talk to the camera, so it can be controlled. That happened some weeks ago and I’ll spare you further details.
Next, I needed a script that would run what I call alternating time lapse. That is acquire a frame after a short interval (60 sec), then after a long interval (4 min), then the short interval, then the long, and so on for three hours. At Nottingham, Darren wrote one for me, using LabView, a rather expensive piece of software that I don’t have her. But Micro-Manager has a scripting language and thanks to help from one of the MM doyens, Chris Weisiger I have a script that will do alternating time lapse. Let the experiments begin!
The first was to see if roots grown here have an oscillation. There would be no point in seeing whether growth on top of the agar, or without a shoot, quenched the oscillation if there was no oscillation in the first place. Maybe it is a UK thing?
My student Xiaoli took the 3-hour movie (having that kind time is lost when sabbatical ends), and I crunched it, only to find some of the velocity profiles were crap. In troubleshooting this, I noticed that between some pairs of frames the light intensity was unstable. The bulb was flickering, or so it seemed. In doing some tests the next day, we found that, with 30 sec between frames, the velocity profiles were fine. This seems odd because the frequency of visible lamp flickers is higher than 30 sec. Nevertheless, needs must and Xiaoli shot another 3 h sequence, this time with 30 sec between frame pairs rather than the usual 60 sec. Happily all the velocity profiles were fine and even more, there was an oscillation (figure 2). Hooray!
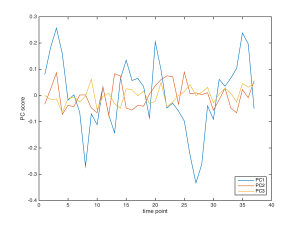
Clearly, PC1 has a substantial oscillation and it is about the same magnitude as I saw in UK. While this is immensely reassuring to see, phenomena that occur in the UK only being perhaps of limited interest, there is a worry: the period is faster than any root I imaged over there. Well, sure, the period could reflect some random difference in growth conditions. But I am worried because the period looks more or less doubled. Could this reflect the frame interval’s being halved? Logically, I cannot see a connection, but I don’t like coincidences.
That is where I am to date. What happens next is solving the intensity instability so that I can use a 60 sec interval. Even if in principle, a 30 sec interval would provide a sharper time base for this kind of study, it is essential first to rule out any funny business about the velocity profile processing. Fortunately, the metal armature that holds the microscope on its side has a gap right beneath the bulb port. This was no accident – we designed it that way to change the bulb when it burned out. When the port is open, and the bulb removed, another light can be shone through. In particular, I have an IR LED source that should be rock solid. If intensity still flickers then it is a camera problem. If not, I can use the IR for new sequences, with 60 sec intervals, and see what happens. Stay tuned!